Lee Lab
 Choogon Lee, Ph.D.
Choogon Lee, Ph.D.
Florida State University
College of Medicine
Dept. of Biomedical Sciences
1115 West Call Street
Tallahassee, FL 32306-4300
Office: (850) 645-1478, MSR 2300-L
Lab: (850) 645-1508, MSR 2310-N
Dr. Lee's Faculty Profile
Research Interests
We have research projects in two different areas: circadian biology and metabolism.
1. We study the molecular mechanism of the circadian clock using the mouse as our model system. Our circadian clock is our body’s way of telling time, and it drives daily oscillations across our physiology and behavior: not only when we sleep and wake, but also our energy metabolism, digestive function, cardiac function, cognitive and emotional state, and tissue repair. Numerous studies demonstrated that serious diseases such as sleep disorders, diabetes and cancer can arise in humans when the clock is compromised by genetic or environmental factors.
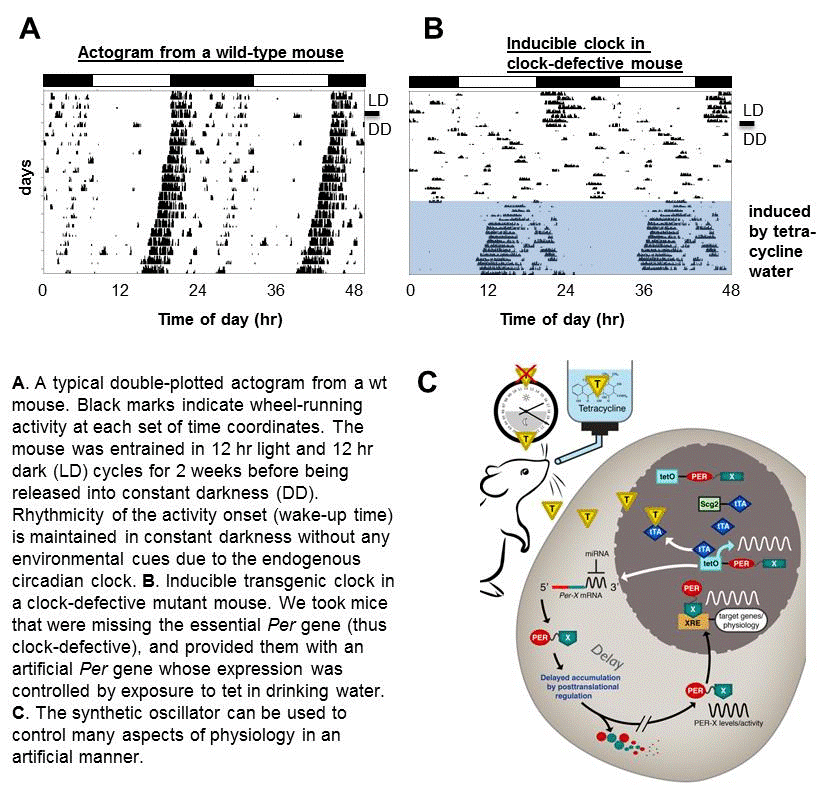
Circadian clocks are found in most organisms, as diverse as cyanobacteria, plants, Neurospora (bread mold), Drosophila, and mammals. In all organisms, the core principle of the clock mechanism is the same: a transcriptional negative feedback loop, consisting of positive elements and negative elements, results in cyclic gene expression with a period of about 24 hours. In mammalian clock cells, CLOCK and BMAL1 are the positive elements, activating transcription of many downstream genes including the negative elements, Period (Per) and Cryptochrome (Cry), whose products form an inhibitory complex. The PER:CRY complex closes the negative feedback loop by inhibiting the positive complex through direct physical interaction. We use mutant and transgenic mouse models along with mathematical models to study how the molecular clock generates time cues for behavior and physiology. For example, we are trying to understand how our wake-up or sleep onset time—measured using behavioral records, “actograms” of wheel running activity—is determined by the clock or disrupted when the clock is compromised, and how the clock directs the expression of important metabolic enzymes when they are needed, around mealtime. Our studies aim to uncover how the clock is regulated normally and to inform how clock dysfunction contributes to pathological states.
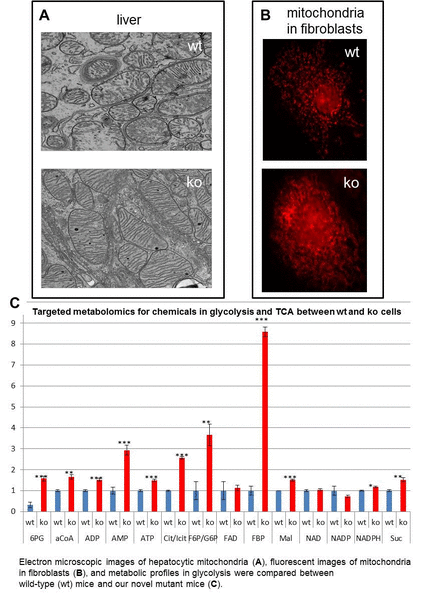
2. We are also studying the regulation and dysregulation of energy metabolism. Serious metabolic diseases such as obesity and type 2 diabetes continue to become more and more prevalent in Western nations due to changes in our dietary habits and lifestyle. While others are trying to tackle this problem through social, psychological, and neurobiological (e.g., modulation of appetite) approaches, we are exploring the possibility of modulating cell-intrinsic metabolic pathways such as glycolysis. We recently discovered serendipitously that mice can lose weight dramatically if the glycolysis pathway is upregulated through modulation of a key enzyme. Our hypothesis is that moderate activation of glycolysis can consume glucose rapidly to the level that can mobilize and burn intracellular and extracellular fat without serious side effects. To study this hypothesis, we plan to use a multi-pronged approach, including generation of transgenic mice overexpressing the target enzyme in an inducible manner, metabolomics, and screening small molecules for allosteric agonists for the target enzyme.
Current Projects
1. Characterization of novel mutant mice for circadian phenotypes: We generate our own novel mutant and transgenic mice or obtain existing mutant mice that were generated and characterized in a non-circadian field based on our hypothesis or preliminary experiments. We measure a full array of circadian parameters from these mice: wheel running activity rhythms, protein and mRNA rhythms of the essential clock genes, coimmunoprecipitation, transcriptional reporter assays, and immunocytochemistry. We are currently characterizing two mutant mice to reveal new molecular mechanisms in clock regulation.
2. Generation of a dynamic 3-D model clock in an artificial cell using mathematical algorithms: In silico approaches are becoming essential to understand the complicated molecular pathways and provide novel insights into developing new research projects leading to effective drug treatments. This is only true when biological variables and parameters are close to the endogenous counterparts. We are measuring important parameters of clock proteins, parameters such as stability, subcellular trafficking, degradation rate and protein-protein interaction in live cells at a single-cell level. We are generating fluorescence-labelled clock proteins, will express them in an inducible manner and monitor them from synthesis to degradation in live cells. Using the measured parameters, we will generate equations and execute them to generate active 3-D images. In both live cells and in silico, we can disrupt genes of interest and see how dynamics of the fluorescence and 3-D images are affected.
3. Regulation of glycolysis activity: We are assessing how upregulation of glycolysis can affect general cell metabolism and whole body physiology. We are using transgenic approaches but will eventually screen small drug compounds to modulate the pathway. We plan to do this transgenic and pharmacological modulation in a simple fat cell model and in a mouse model. Depending on these preclinical studies, we will decide if we will move on to human studies.
Current Lab Personnel
Postdoctoral associate: Yuanhu Jin, PhD
Graduate Student: Kwangjun Lee, MS
Visiting Student: Hyunjeong Joo
Recent Publications
2017
D'Alessandro, M., Beesley, S., Kim, J., Jones, Z., Chen, R., Wi, J., Kyle, K., Vera, D., Pagano, M., Nowakowski, R. & Lee, C. (2017). Stability of Wake-Sleep Cycles Requires Robust Degradation of the PERIOD Protein. Current Biology, 27, 3454-3467. link
Yoo SH, Kojima S, Shimomura K, , Koike N, Buhr ED, Furukawa T, Ko CH, Gloston G, Ayoub C, Nohara K, , Reyes BA, Tsuchiya Y, Yoo OJ, Yagita K, Lee, C., Chen Z, Yamazaki S, Green CB, & Takahashi JS. (2017). Period2 3'-UTR and microRNA-24 regulate circadian rhythms by repressing PERIOD2 protein accumulation. PNAS, 114, E8855-8864. link
2016
Pramudya, I., Rico, C., Lee, C., & Chung, H. (2016). POSS-Containing Bioinspired Adhesives with Enhanced Mechanical and Optical Properties for Biomedical Applications. Biomacromolecules, 17, 3853-3862. doi:10.1021/acs.biomac.6b00805 link
Kettner, N. M., Voicu, H., Finegold, M. J., Coarfa, C., Sreekumar, A., Putluri, N., Katchy, C. A., Lee, C., Moore, D. D., & Fu, L. (2016). Circadian Homeostasis of Liver Metabolism Suppresses Hepatocarcinogenesis. Cancer Cell, 30(6), 909-924. doi:10.1016/j.ccell.2016.10.007 link
Matsu-Ura, T., Dovzhenok, A., Aihara, E., Rood, J., Le, H., Ren, Y., Rosselot, A. E., Zhang, T., Lee, C., Obrietan, K., Montrose, M. H., Lim, S., Moore, S. R., & Hong, C. I. (2016). Intercellular Coupling of the Cell Cycle and Circadian Clock in Adult Stem Cell Culture. Mol Cell, 64(5), 900-912. doi:10.1016/j.molcel.2016.10.015 link
2015
D’Alessandro, M., Beesley, S., Kim, J.K., Chen, R., Abich, E., Cheng, W.,Yi, P., Takahashi, J.S., Lee, C. Circadian rhythms can be generated and modulated by an artificial circadian clock in clock-defective mammals. Nature Communications, accepted. link
Kettner, NM, Mayo, SA, Hua J, Lee, C, Moore, DD, Fu, L. (2015) Circadian Dysfunction Induces Leptin Resistance in Mice. Cell Metab 22: 1-12. link
Cao, R., Gkogkas, CG., de Zavalia, N., Blum, ID., Yanagiya, A., Tsukumo, Y., Xu, H., Lee, C., Storch, K., Liu, AC., Amir, S. & Sonenberg, N. (2015) Light-regulated translational control of circadian behavior by eIF4E phosphorylation. Nature Neuroscience 18: 855-62 link
2014
Penas, C., Ramachandran, V., Simanski, S., Lee, C., Madoux, F., Rahaim, R.J., Chauhan, R., Barnaby, O., Schurer, S., Hodder, P., Steen, J., Roush, W.R., Ayad, N.G. (2014) Casein Kinase 1δ Dependent Wee1 Degradation. JBC 289: 18893-903 link
2013
Chen, R., Dalessandro, M., Lee, C. (2013) miRNAs are required for generating a time-delay critical for the circadian oscillator. Current Biology 23: 1959 link
Bhatwadekar, A., Yan, Y., Qi, X., Thinschmidt, J., Neu, M., Calzi, S., Shaw, L., Dominguez, J., Busik, J., Lee, C., Boulton, M., Grant, M. (2013) Diabetes 62: 273 link
2012
Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. (2012) Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science 338: 349 link
2011
Lee, H., Chen, R., Etchegaray, J.P., Weaver, D.R., Lee, C. (2011) The balance between CK1 and PP1 regulates PER phosphorylation and circadian oscillator speed. PNAS 108: 16451-6 PMCID: PMC3182690 link
Lee, Y., Chen, R., Lee, H., Lee, C. (2011) Stoichiometric interaction among clock proteins regulates robustness of circadian rhythms. JBC 286: 7033-42 link
2010
Lee, C., Chen, R., Lee, H. (2010) PERpetual motion of the circadian feedback loop. Cell Cycle 9: 853-4. link
2009
Lee, H., Chen, R., Lee, Y., Yoo, S., Lee, C. (2009) Essential roles of CKId and CKIe in the mammalian circadian clock. Proceedings of the National Academy of Sciences (PNAS)106: 21359.
Chen, R., Schirmer, A., Lee, Y., Lee, H., Kumar, V., Yoo, S., Takahashi, J.S., Lee, C. (2009) Rhythmic mPER abundance defines a critical nodal point for negative feedback within the circadian clock mechanism. Molecular Cell 36: 417-430 link
Ramsey, K.M., Yoshino, J., Brace, C.S., Abrassart, D., Kobayashi, Y., Marcheva, B., Hong, H., Chong, J.L., Buhr, E.D., Lee, C., Takahashi, J.S. and Imai, S., Bass, J. (2009) Circadian Clock Feedback Cycle Through NAMPT-Mediated NAD+ Biosynthesis. Science 324:651. link
Ansari, N., Agathagelidis, M., Lee, C., Korf, H., von Gall, C. (2009) Differential
maturation of circadian rhythms in clock gene proteins in the suprachiasmatic nucleus and the pars tuberalis during mouse ontogeny. European Journal of Neuroscience. 29:477-89 link
