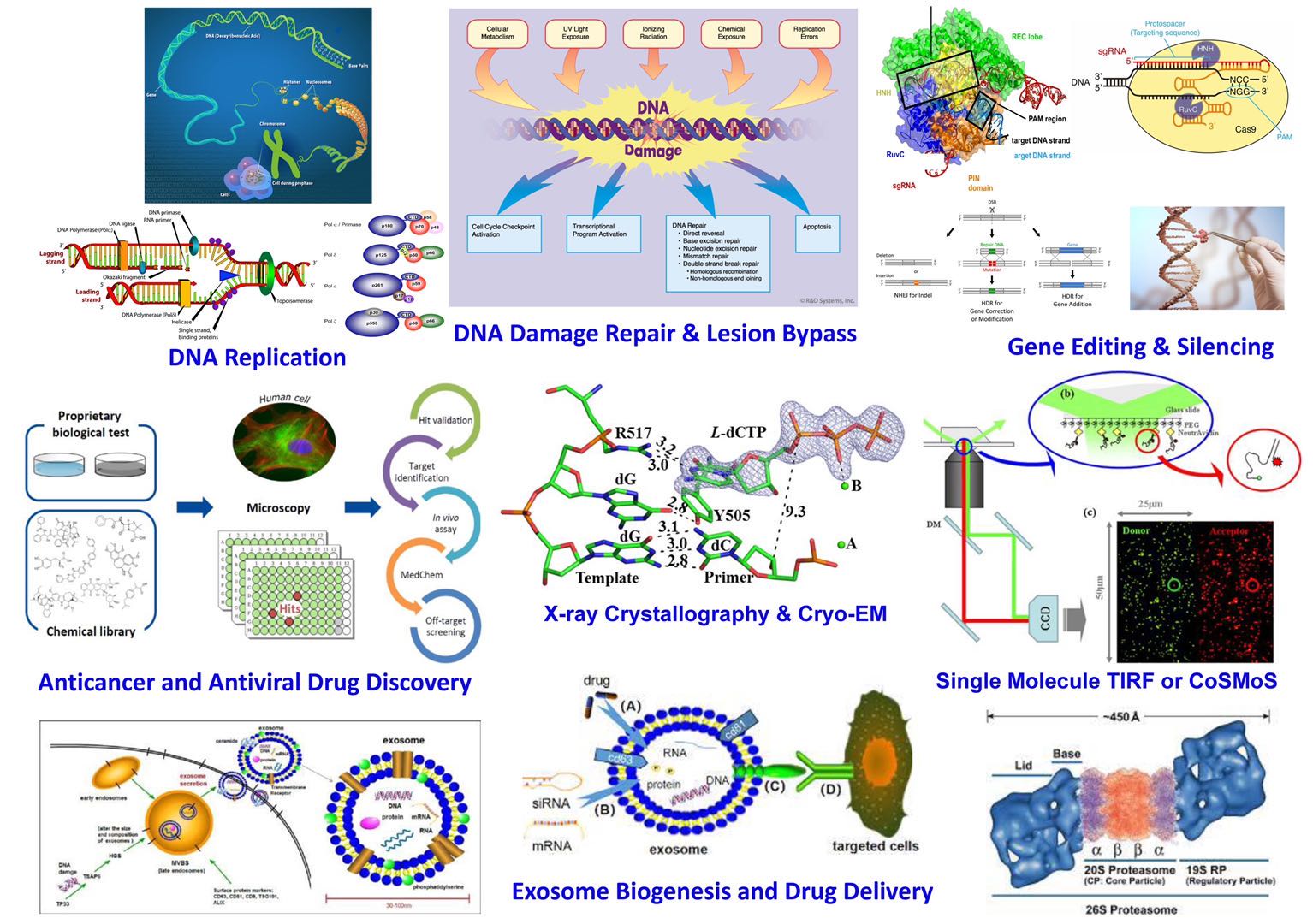
Research Interests
The Suo Laboratory is a biochemistry, biophysics and molecular biology laboratory with focus on advanced enzymology, structural biology, and drug design and delivery. We elucidate mechanisms of DNA/RNA replication, DNA lesion bypass, and DNA base excision repair catalyzed by various human, archaeal, and viral DNA/RNA polymerases. We also mechanistically investigate gene editing enzymes, e.g. CRISPR-Cas9 and Cas13a, and yeast proteasome. The Suo Laboratory utilizes a variety of techniques to investigate intriguing questions in enzymology and to pursue rational drug design and drug delivery. Pre-steady-state kinetic methods including rapid chemical quench-flow and stopped-flow allow us to follow enzymatic reactions in the millisecond time scale and to extract more kinetic information than the traditional steady-state kinetic methods. We also use single molecule spectroscopic methods including smFRET and CoSMoS to investigate the dynamics of individual protein molecules and assembly kinetics of large protein complexes which are difficult to be analyzed through ensemble experiments. X-ray crystallography and single particle Cryo-EM are being used to solve the atomic resolution structures of small and large enzyme-substrate complexes. These multi-disciplinary approaches will allow us to develop new mechanistic methods and to advance enzymology into unprecedented territory. Our understanding of the kinetics, dynamics, and structure of enzyme targets is used for rational antiviral and anticancer drug design. The designed enzyme inhibitors are being chemically synthesized and tested in cell- and animal-based models. Since 2019, the Suo Laboratory has been using extracellular vehicles and liposomes to deliver potent small and large molecule inhibitors to treat various diseases. To achieve these goals, the Suo Laboratory has been recruiting highly dedicated and motivated undergraduate and graduate students, postdoctoral researchers, and staff scientists to carry out the following research projects:
1. Biology and utilization of loaded extracellular vesicles and liposomes in disease treatment
Exosomes (small extracellular vesicles) are extracellular membrane-enclosed vesicles with size ranging from 30 to 200 nm in diameter secreted by most types of human cells, which are formed inside the multivesicular bodies and thus consist of many macromolecules from their parent cells. Various proteins, receptors, various enzymes, transcription factors, lipids, nucleic acids (microRNA and mRNA), and extracellular matrix proteins have been identified in exosomes. Interestingly, some proteins specifically arise from parental tissues while others are common among all exosomes. Due to unique properties, mesenchymal stem cells (MSCs) and hematopoietic stem cells (HSCs) have been used in various therapies for wound healing and injured tissue regeneration. In placental mammals, an umbilical cord connects a placenta to a developing embryo. Umbilical cords are rich source of MSCs and HSCs and such MSCs have been used to treat different types of disorders like bone repair, neurological diseases, cancer, cardiac and liver diseases. Interestingly, exosomes secreted by MSCs can replace stem cell-based therapies in various wound-healing models. Exosomes have also been used as drug delivery vehicles for non-native therapeutics such as nucleic acids and small drug molecules. The Suo Laboratory is exploring the usage of exosomes to deliver therapeutic molecules for wound healing and for the treatment of different forms of cancers, cytokine storm, and other diseases.
2. Genome editing through CRISPR-Cas9
Using a family of DNA sequences, named Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR), in conjunction with CRISPR-associated (Cas) proteins (RNA-guided endonucleases), numerous bacteria and archaea can recognize non-native genetic material and target it for digestion in order to prevent viral infections. The CRISPR-Cas systems serve as an adaptive immunity to protect bacteria and archaea from viruses as well as regulate the incorporation of plasmids from other prokaryotes. The discovery of such a bacterial adaptive immune system and associated programmable endonucleases has revolutionized genomic editing. A simple CRISPR-Cas system from Streptococcus pyogenes, relying on a single Cas9 protein (an endonuclease) and two small RNA molecules, crRNA and trans-activating CRISPR RNA (tracrRNA), has rapidly become one of the most useful tools for editing genomes. Guided by a programmable RNA sequence, the Cas9 endonuclease cuts both strands of DNA at a target sequence. Once the DNA is cut, cellular DNA repair machinery will add or delete pieces of genetic material, or insert a customized DNA sequence through a donor DNA plasmid. Due to its ease of use and simple programming platform, the CRISPR-Cas9 system has a wide variety of applications including basic biological research, development of biotechnology products, and treatment of diseases. The Suo Laboratory focuses on understanding the mechanisms underlying the Cas9 functions by using a variety of cutting-edge methods such as pre-steady-state kinetics, single molecule fluorescence microscopy, X-ray crystallography, and Cryo-EM.
3. DNA base excision repair
Cellular genomic integrity is constantly damaged by endogenous and exogenous processes and agents, for example, oxidation, hydrolysis, and exposure to radiation and chemicals. As a result of these factors, cellular DNA contains many different forms of chemical alterations, called DNA lesions. It is estimated that each mammalian cell has to repair about one million DNA lesions each day. If unrepaired, DNA lesions will either cause cell death or gene mutations. To avoid these adversities, cells have evolved several effective DNA repair pathways including base excision repair (BER). BER specializes in repairing single base lesions including oxidized, alkylated, or methylated bases. To replace a lesion with a natural base, BER recruits four enzymes and a protein cofactor to perform five sequential enzymatic activities: base excision by a glycosylase, single-strand DNA cleavage by an endonuclease, DNA synthesis by a polymerase, excision of a deoxyribose and its 5¢-phosphate by a lyase, and finally the ligation of nicked DNA by a ligase. The Suo Laboratory investigates the mechanisms of individual BER enzymes through pre-steady-state kinetic methods and single molecule fluorescence techniques. We also employ X-ray crystallography and Cryo-EM to solve the structures of individual enzymes and protein complexes of BER.
4. Enzymology of yeast proteasome assembly and function
The proteasome is the largest and most complicated protease in eukaryotes, and is deregulated in numerous human diseases. The proteasome consists of almost 70 individual subunits that must precisely assemble in defined orders and three-dimensional arrangements to yield mature, functional particles. To understand proteasome assembly and function, we are collaborating with Prof. Robert Tomko’s Laboratory within the same Department to study yeast proteasome by employing single molecule techniques (smFRET and CoSMoS) and ensemble methodologies (stopped-flow FRET and pre-steady-state kinetics).
5. Kinetic, dynamics, and structural investigation of DNA lesion bypass catalyzed by Y-family DNA polymerase
DNA lesions often block DNA replication, so cells possess specific, often error-prone, DNA polymerases to bypass such lesions and to promote replication of damaged DNA. More than 220 DNA lesion bypass polymerases have been discovered. Most of these polymerases, which share sequence similarity and catalyze DNA polymerization with low fidelity and poor processivity, are classified into the Y-family. For example, Dpo4 is a lone Y-family polymerase from Sulfolobus solfataricus, a hyperthermophilic archaeon. Human polymerases eta (η), iota (ι), kappa (κ), and Rev1 are examples of eukaryotic DNA lesion bypass Y-family enzymes. Pol η, encoded by hRAD30A, bypasses cis-syn pyrimidine-pyrimidine dimers efficiently and accurately. Mutations in hRAD30A inactivate Pol η and lead to UV-induced mutagenesis and skin cancer. During lesion bypass, a Y-family polymerase will be switched onto damaged DNA while a replicative polymerase is blocked and switched off from the lesion site. The Suo Laboratory is using pre-steady-state kinetic methods, single molecular techniques, X-ray crystallography, and Cryo-EM to decipher the detailed mechanisms of lesion bypass catalyzed by Dpo4 or a human Y-family polymerase. We have developed a novel high-throughput short oligonucleotide sequencing assay (SOSA) to determine the DNA sequence of lesion bypass products synthesized by the Y-family enzymes. A better knowledge of the Y-family polymerases based on our results will facilitate the understanding of cancer formation.
6. Mechanistic and structural studies of human DNA polymerase holoenzymes
During eukaryotic DNA replication, three polymerases (α, δ, and ε) participate in the synthesis of the leading and lagging strands. High-fidelity DNA replication is essential to genomic stability and is mainly contributed by the action of DNA polymerases ε and δ. Each of these replicative polymerases is a holoenzyme of one large catalytic subunit and three small subunits with both polymerase and exonucleolytic proofreading activities. So far, hundreds of cancer-associated mutations have been identified in the genes of DNA polymerases ε and δ. Our research aims to employ pre-steady-state kinetics, single molecule fluorescence techniques, X-ray crystallography, and Cryo-EM to elucidate the structural and kinetic basis of these mutations that drive mutagenesis and tumor development.
7. Investigation of SARS-CoV-2 RNA-dependent RNA polymerase and develop small molecule inhibitors
A novel coronavirus (SARS-CoV-2) has caused a global pandemic resulting in almost three million infections and close to 200,000 deaths worldwide. SARS-CoV-2 replicates its RNA genome through a virally encoded RNA-dependent RNA polymerase (RdRp), a major target for developing small molecule drugs to treat COVID-19. For example, remdesivir, a promising nucleotide analog inhibitor of SARS-CoV-2 RdRp, is in several Phase III clinical trials. The Suo Laboratory is mechanistically investigating SARS-CoV-2 RdRp. We will also screen for potent SARS-CoV-2 RdRp inhibitors from our nucleotide analog library. If we find such hit compounds, we will perform SAR to optimize the hits and develop lead compounds. Subsequently, we will collaborate with the CDC to test our lead inhibitors in their cell- and animal-based models.