Research
Research Interests
Sleep and circadian rhythms are crucial aspects of our physiology and behavior and are involved in or affected by many disease states. The mission of our lab is to advance our understanding of the molecular mechanisms underpinning these processes and their disorders. In doing so, we will provide important insights for treating a wide variety of diseases in which sleep disorders are major causes, symptoms, or comorbidities.
Sleep is regulated by two distinct but interconnected mechanisms: sleep homeostasis and the circadian clock. Optimal sleep quality requires alignment of these two pathways, and disruptions in either one can affect the other and lead to sleep disorders. It has been hypothesized that the multiple brain centers forming the homeostatic mechanism regulate the total amount of sleep and sleep depth, while the suprachiasmatic nucleus (SCN) circadian clock regulates timing of sleep/wake onset and continuous sleep. In the last 30 years, great strides have been made in elucidating the molecular mechanisms of the circadian clock, but there remains much that is unknown, and the molecular basis for sleep homeostasis and the crosstalk between the clock and the homeostat remain almost entirely mysterious. Our lab uses a mix of mouse and cell-based models to both deepen our molecular understanding of the circadian clockwork as well as shine a light on the mechanisms of sleep homeostasis.
Current Projects
We are pursuing three lines of research toward the overarching goal.
1) The genetic basis of human chronotypes and sleep disorders: Despite a clear causal link between genetic polymorphism in clock genes and sleep disorders, systematic investigation of pathogenic mutations in human clock genes has not been conducted for two reasons: The first is the abundance of natural genetic polymorphisms in human clock genes. There are thousands of known polymorphisms in major clock genes, a subset of which may affect chronotypes and sleep disorders. The second is the historical reliance on rodent models for functional assessment, which are time- and resource-intensive. To address this gap, we developed a cell-based in vivo-like platform that allows us to screen and characterize many mutations in a cost- and labor-effective manner (Figure 1). Using our system, we have to date assessed dozens of natural human polymorphisms as well as hypothesis-based mutations in known clock genes, and we have identified novel mechanisms and mutations that can cause dramatically altered rhythms and sleep cycles (Figure 2). During the course of these analyses, we found common themes that allow us to identify potential pathogenic mutations from the existing thousands of natural human mutations in clock genes and predict their phenotypes.
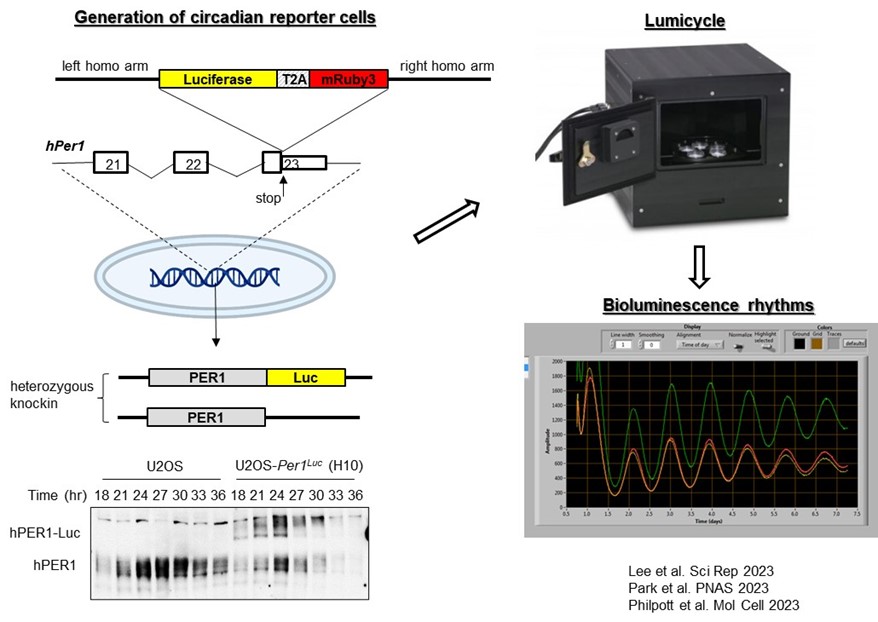
Figure 1. Circadian rhythms can be accurately measured by a luciferase endogenous reporter. Firefly luciferase along with a fluorescent protein can be knocked into clock genes for real-time reporting of circadian rhythms. In this case, the dual reporter was knocked into the human Per1 gene in U2OS cells using CRISPR. As shown in this case, the majority of knockins were heterozygotes. The knockin did not disrupt the function of the Per1 gene significantly as evidenced by similar protein oscillations of PER1-Luc compared to the wild-type (wt) allele. When bioluminescence was measured from these cells using a Lumicycle bioluminometer, they showed robust rhythms which were modulated by significant polymorphic mutations in clock genes.
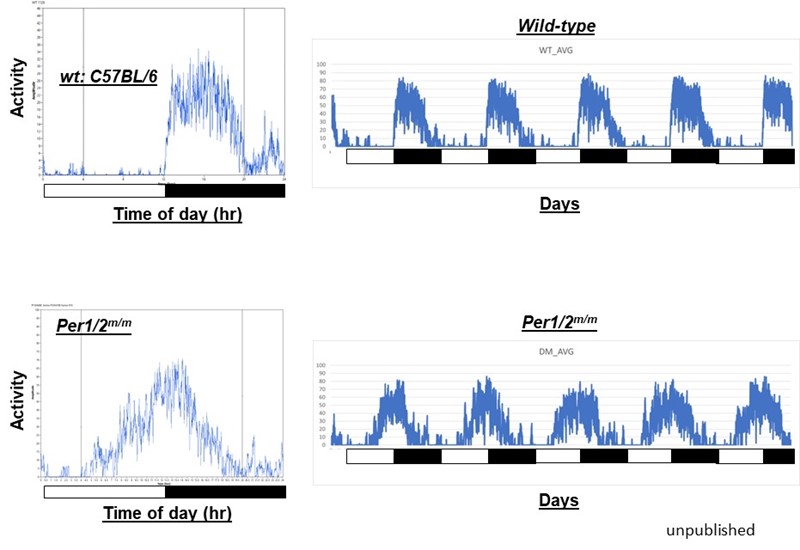
Figure 2. Sleep cycles are dramatically altered by polymorphic mutations in Per genes. These mutations (SNPs) were identified using our reporter cells. Two activity graphs are shown: one from a wt mouse (top) and the other from a mutant mouse (bottom) harboring point mutations in Per1 and Per2 genes at a homologous location. The white and black bars represent 12 hr light and 12 hr dark cycle, respectively. The wt mouse shows a typical nocturnal behavior while the mutant mouse exhibits half diurnal and half nocturnal behavior. It is expected that a variety of point mutations in this region will also produce this kind of advanced wake/sleep cycles in humans.
2) The molecular link between the circadian clock and sleep homeostasis: Sleep research is a largely uncharted territory at the molecular level. To date, no genes or molecular pathways have been shown definitively to be critical for sleep homeostasis. To understand how our sleep homeostasis is regulated at the molecular level and modulate our sleep to our advantage, it would be imperative to identify major sleep genes and pathways.
As discussed above, the sleep homeostasis is distinct from the circadian clock. However, there must be crosstalk between the two pathways. The circadian clock must be able to measure or regulate temporal sleep pressure so that sleep or wake onset can occur at the same circadian time after sleep pressure builds to a certain threshold or is relieved to a basal level (Figure 3). The presence of the crosstalk is further supported by altered timing and duration of sleep in circadian mutant animals with altered circadian periods. However, the molecular basis for this link has largely been unexplored in both circadian and sleep research.
Recently, we serendipitously found that a Per2-driven clock in Per1 knockout (KO) mice measures or regulates temporal sleep pressure, in addition to driving circadian rhythms. It seems that the Per2 oscillator can gauge the level of sleep pressure at different circadian times, and the oscillator can be reset to CT12 (activity onset and circadian time with the lowest sleep pressure) from any time point during the active phase by long light pulses. We believe sleep pressure is reset to the basal level because the Per2 clock directly controls the sleep pressure as it is reset to a specific circadian time point CT12. However, in wt mice, we believe the Per1 oscillator maintains circadian momentum and resists phase shifts under these conditions. We are following this lead with the goal to identify genes, molecules and neurochemicals that link the clock to the sleep homeostasis using the clues we will obtain from the behavioral experiments. We expect the levels of sleep pressure-linked molecules to follow a specific circadian rhythm that is modified by these conditions. We would screen for these molecules from circadian brain samples from Per1 KO mice by diverse techniques such as HPLC/Mass spec.
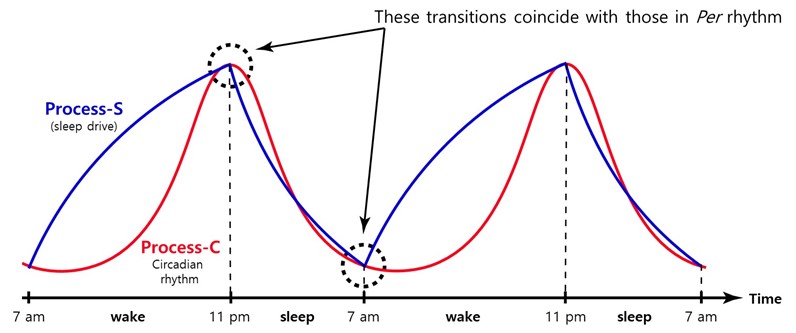
Figure 3. Two-process model for sleep regulation. Little is known about how the circadian clock is integrated with the sleep drive to regulate wake or sleep timing. This conceptual model posits that sleep pressure or drive increases as soon as we are awake and dissipates gradually after we fall asleep, and the timing of transitions is controlled by the clock. The circadian rhythm is represented by the pacemaker Per rhythm in the master clock tissue SCN. However, our data suggest that in Per1 KO mice, these two processes are jointly regulated by the Per2 oscillator, which provides us with a unique opportunity to dissect the sleep homeostasis pathway at the molecular level.
3) Identification of novel clock genes: The human cell line U2OS is known to possess a functioning circadian clock and is widely used as a model system. We engineered this cell line to express fluorescent protein-fused clock proteins from endogenous loci and thus report expression of endogenous clock genes in living cells. We have shown that this novel cell platform can be combined with powerful CRISPR lentiviral libraries targeting all human genes (e.g. GeCKO v2), as well as FACS sorting and high-throughput sequencing methods, to identify novel candidate clock genes that alter expression of the reporter genes. In Figure 4, when our Bmal1-mRuby3 reporter cell line was infected with the library lentivirus, some cells expressed significantly higher mRuby3 signals; these cells were selected and enriched by FACS as shown in the bottom graph. The corresponding sgRNAs for the CRISPR KO genes will be PCR-amplified and sequenced and ranked by frequency.
We are refining this methodology by developing cells where fluorescent protein mRuby3 (or mClover3) in tandem with Luciferase (Luc), separated by the cleavage element T2A, is knocked into human clock genes Per1, Per2, Bmal1, and Cry1. The fluorescent proteins will remain attached to the clock proteins while the cleaved Luc will provide a circadian bioluminescence rhythm. Our hypothesis is that knocking out candidate clock genes will change reporter protein levels (which correspond to endogenous clock protein levels) whether the novel gene acts transcriptionally or posttranscriptionally. We will then validate the circadian phenotype by measuring the Luc rhythm in isolated clones. This platform technology can be used for identifying new genes in any biological pathways and drug screening.
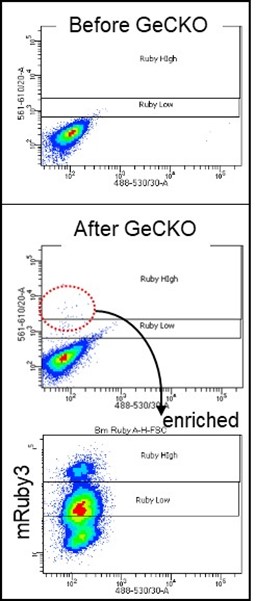
Figure 4. BMAL1-mRUBY3 levels are elevated in some of the GeCKO lentivirus-infected cells. We generated a GeCKO v2 human lentiviral library and tested its integrity by random sequencing. We adjusted infection titer into our reporter cells so that only 1-2 viral copies can be integrated into the genome by serial dilution and digital PCR. Note that there are two groups of cells above the control reporter cell after enrichment of initially selected cells. Once candidate genes are identified by NGS, reverse genetics will be performed to confirm the original data and study how the clock is regulated by these genes.
Current Funding
Pending
NIH/NIGMS
Project Number: R35 MIRA (PI: Choogon Lee), Impact score: 25
Period: 12/1/2024-11/30/2029
Title: Genetic basis of circadian rhythms and sleep disorders
Description: This pending MIRA application overlaps with the two active NIGMS awards and will replace them if funded.
Active
NIH/NIGMS
Project Number: 1R01GM131283-01A1 (PI: Choogon Lee)
Period: 9/4/19-8/31/2025, NCE
Title: Molecular mechanisms underlying human circadian sleep disorders
Description: The overall goal of this application is to study how mutations in Period genes can cause sleep disorders and to develop mouse models carrying mutant Period genes.
NIH/NIGMS
Project Number: GM147340-01A1 (PI: Choogon Lee)
Period: 8/23/2023-6/30/2027
Title: A novel cell-based platform to study human circadian disorders
Description: The overall goal of this application is to develop cell-based systems to test functionality and pathogenicity of human SNPs in major clock genes, CK1, Bmal1 and Clock.
FL Department of Health
Project Number: Ed and Ethel Moore AD Research Program 24A04 (PI: Choogon Lee,
coPI: Aaron Wilber, Samuel Grant)
Period: 3/1/2024-2/28/2027
Title: Targeting the orexin pathway to treat sleep disorders and improve
glymphatic clearance in Alzheimer’s disease
Description: The overall goal of this application is to treat sleep disorders in Alzheimer’s disease rodent models.

