Ubiquitin-dependent proteolysis in health and disease
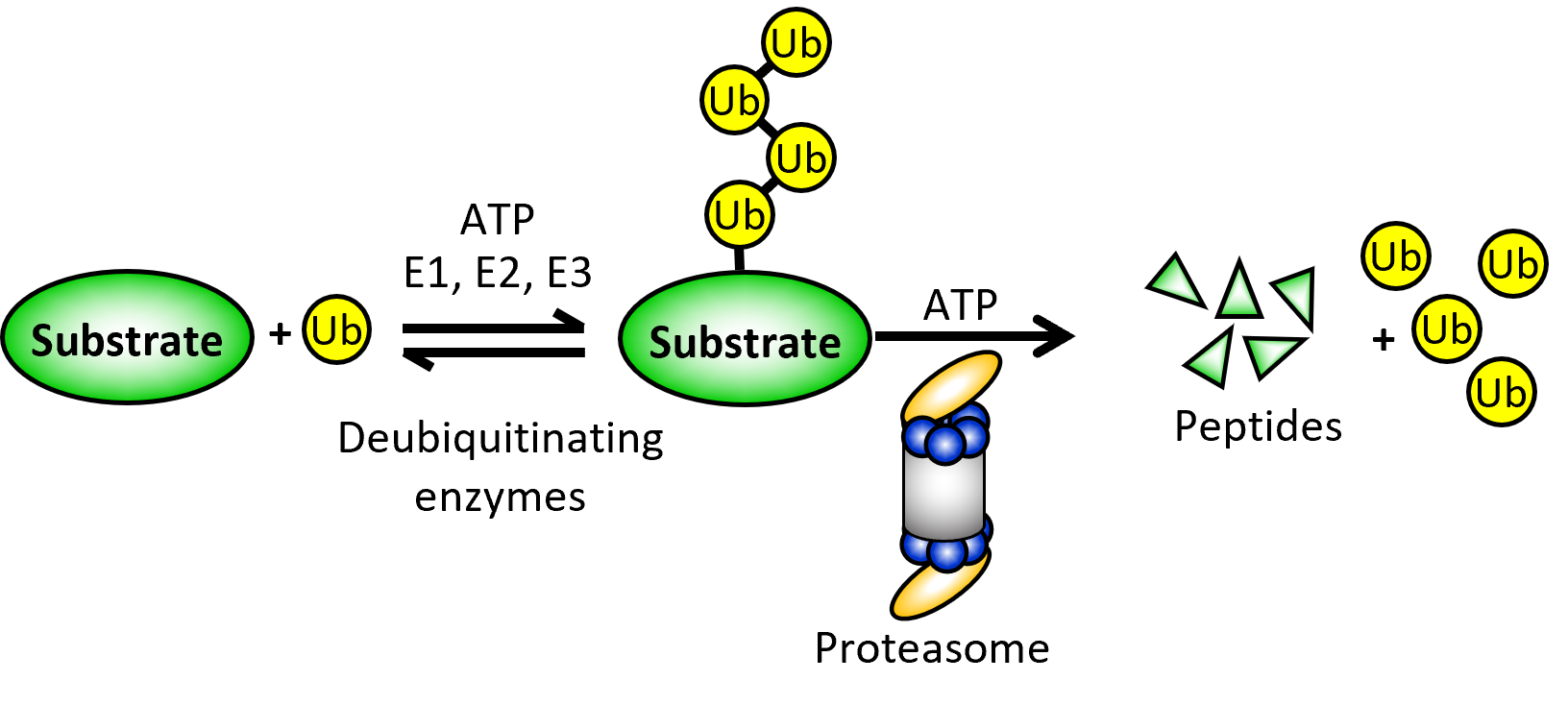
Work in the Tomko lab centers on the ubiquitin-proteasome system (UPS), which mediates ~80% of all regulated (e.g., intentional) protein degradation in all eukaryotes. Our group has traditionally focused on understanding the assembly and function of the 26S proteasome, which is the endpoint for proteins destined for degradation by the UPS. It is also the largest and most complicated protease known. Deregulation of proteasomal proteolysis underlies diverse human diseases, including cancer, neurodegeneration, diabetes, and several autoimmune disorders. By better understanding the structure, biogenesis, and function of the proteasome, we hope to identify new targets for disease treatment and discover small molecules that modulate these targets.
More recently, we have also become interested in the UPS of a family of infectious obligate intracellular parasites known as microsporidia. These fascinating parasites spread easily through contaminated food and water sources, but have largely been ignored by modern medicine because they have traditionally only caused disease in immunocompromised patients. However, as long-term use of immunosuppressive drugs becomes the standard of care for many conditions, including organ transplants, chronic inflammatory diseases, and autoimmune disorders, they are emerging as a new and serious health threat. We are thus characterizing the highly divergent UPS in these parasites in an effort to develop novel therapeutics for this important but virtually ignored class of pathogens.
Proteasome assembly and function
The proteasome consists of ~70 individual protein subunits that must occupy defined positions within the final complex to ensure its efficient function. Further, individual subunits within the proteasome are sometimes substituted with alternative forms to yield specialized proteasomes with altered specificities and activities. These alternative proteasomes are involved in specialized biological processes, such as T-cell selection, generation of epitopes for antibodies, and maturation of sperm cells. Further, changes to the composition of the proteasome occurs frequently in human cancers. Thus, understanding how this unusually large and complicated cellular machine forms and functions is guiding the development of drugs for autoimmunity, cancer, fertility, and infectious disease, among others.
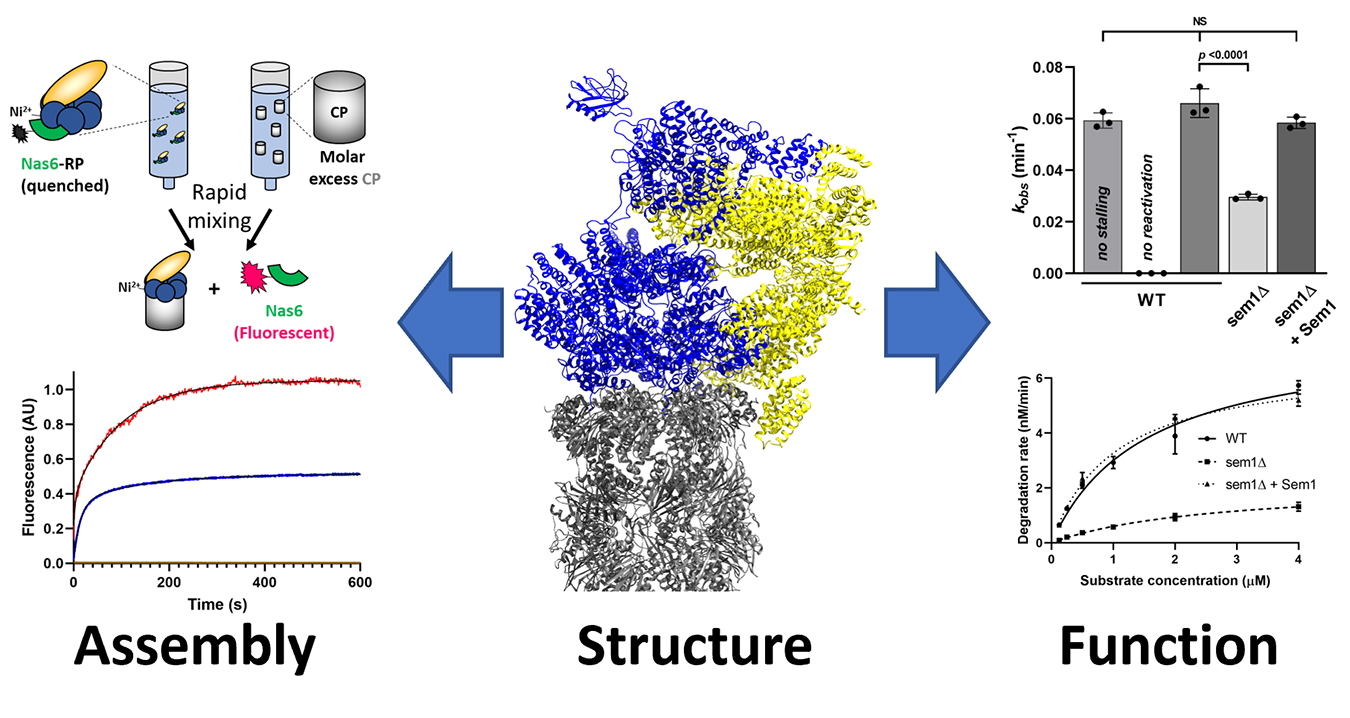
If the subunits of the proteasome were to assemble randomly (e.g., driven solely by stochastic collisions between subunits), there are more possible assembly sequences than there are stars in the known Universe. Surprisingly, only two or three of these assembly sequences seem to occur in the cell. How is this remarkable complexity reduced and managed to assure that a properly composed, functional complex is always the endpoint? How does the cell make specialized proteasomes when needed? We are interested in understanding the molecular and cellular mechanisms that enforce this ordered, hierarchical assembly of the proteasome in vivo. We use approaches pulled from biochemistry, biophysics, protein engineering, chemical genetics, proteomics, and cell biology to better understand this process, with the ultimate goal of identifying bottlenecks in the assembly process that could be enhanced or further constricted to treat human disease.
Although assembly is our main goal, we occasionally stumble upon interesting things that improve our understanding of how the proteasome selects and degrades its substrates. The proteasome contains ~20 individual enzymatic centers, and it processes substrates for degradation almost like an assembly line building a car, but in reverse. Our secondary interest is in understanding how these different processing sites are functionally coupled within the proteasome during catalysis, and how their activities are restrained during proteasome assembly to prevent premature and incomplete substrate processing.
The ubiquitin-proteasome system of microsporidia
Microsporidia possess a highly divergent 26S proteasome compared to all other eukaryotes, and appear to be missing many accessory proteins and assembly chaperones that would normally promote efficient proteasome biogenesis and function. This raises several important questions – for example, why is it that these organisms lack proteins that are essential for survival in all other eukaryotes examined? Can these divergent proteasomes assemble without exogenous assistance from the chaperones required in humans? What are the differences in the structure and function of this divergent proteasome, and how can we target these differences to kill off the parasites without affecting the host cells? We have developed a suite of biochemical tools that will help us begin answering these questions. We anticipate that this divergent proteasome may make for an outstanding drug target for microsporidiosis.
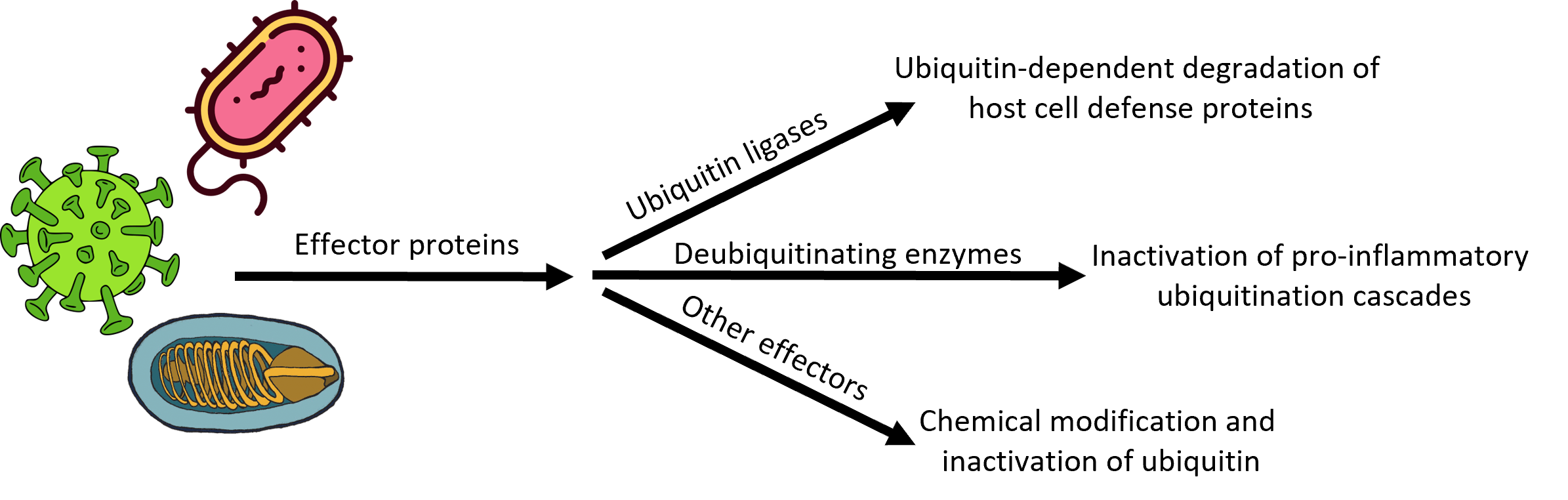
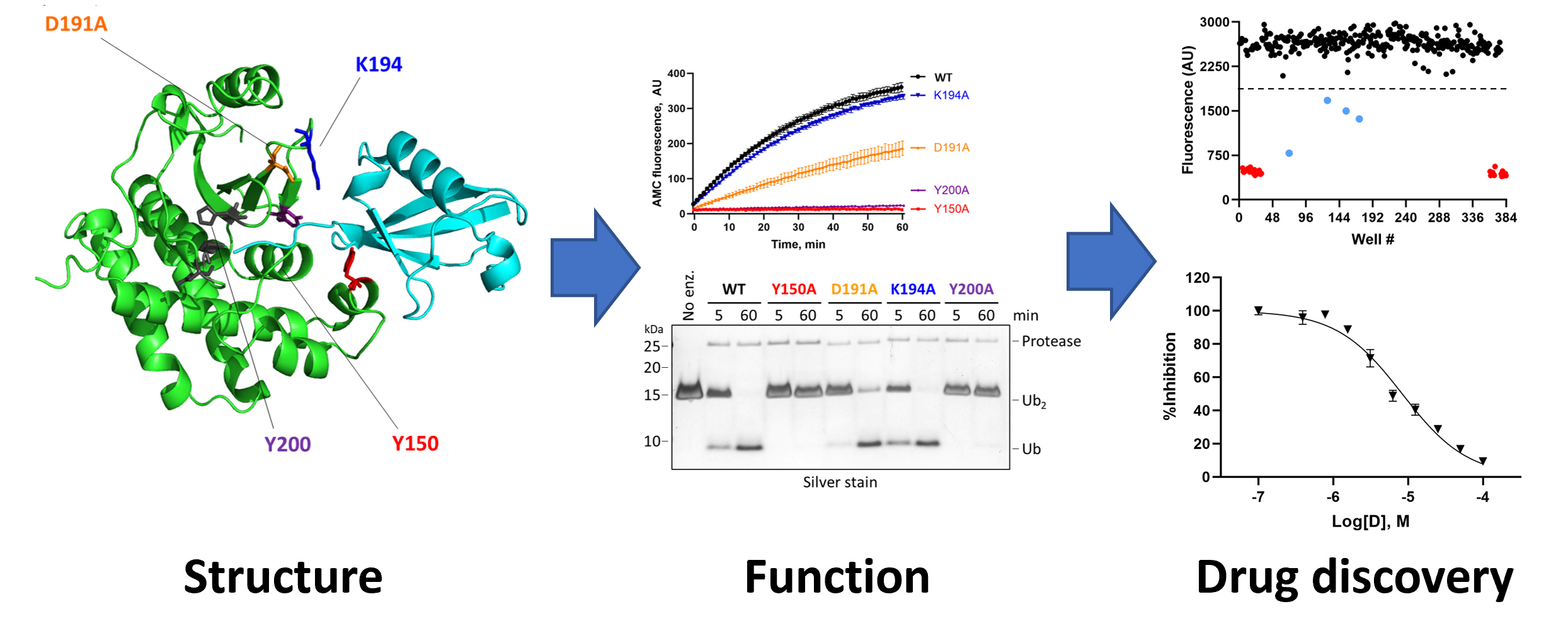
Microsporidia also seem to have an unusual collection of ubiquitin ligases and ubiquitin proteases—including many that are secreted into the infected host cell—whose roles in infection, establishment of an intracellular niche, replication, and dissemination to neighboring cells are completely unknown. We are using biochemical and biophysical approaches coupled with transgenic and cell-based models of microsporidial infection to understand the structure, function, substrate selectivity, and physiological roles of these microsporidial UPS components en route to our larger goals of developing first-in-class drugs for the treatment of microsporidiosis.